Text mining allows for the rapid review and analysis of large volumes of biomedical literature, giving life science companies valuable insights to drive R&D and inform business decisions.
Given the easy accessibility of article abstracts through such databases as MEDLINE, many researchers use this summary information to identify a collection of articles (or “corpus”) for use in text mining rather than taking steps to obtain the full text of the articles. While abstracts provide some valuable pieces of information, there are limitations in using abstracts that can affect the quality of text mining results when compared to the results of mining a corpus of full-text content.
Some in the research community find abstracts to be good enough for their purposes. We’ve heard these defenses for mining abstracts over full-text.:
- “More text means more room for false positives.”
- “Abstracts are more easily accessible via biomedical databases.”
- “We don’t have the time or resources to spend on additional data cleansing and normalization work for unstructured content.”
While there are kernels of truth to each of these challenges, text mining full-text articles over abstracts has significant benefits. Now, new research from bioinformaticians at the University of Copenhagen and the University of Denmark confirm that vital information goes undiscovered when abstracts are mined rather than full-text articles.
Inside the largest comparative study of text mining abstracts vs. full text articles
The study, released this month on bioRxiv, an online archive and distribution service for unpublished preprints in the life sciences, involved the analysis of more than 15 million full-text scientific documents and their abstracts published between 1823 and 2016. These articles, mainly in PDF format, comprised articles published by Elsevier, Springer, and those in the Open-Access subset of PMC.
The team compared their findings from the corpus of full-text articles to the corresponding results from the matching set of abstracts included in MEDLINE.
Here’s a look at some of the report’s main takeaways:
Full text outperformed MEDLINE abstracts in all benchmarked cases
To showcase the potential of text mining full-text articles, the team extracted published protein-protein, disease-gene, and protein subcellular associations using a named entity recognition system.
In every case, the results showed that mining the full-text article corpus outperformed the same analysis using abstracts only.
“Through rigorous benchmarking and comparison of a variety of biologically relevant associations, we have demonstrated that a substantial amount of relevant information is only found in the full body of text,” the report indicates.
This suggestion isn’t the first of its kind. Back in 2010, a study published in the Journal of Biomedical Informatics found that only 8% of the scientific claims made in full-text articles were found in their abstracts.
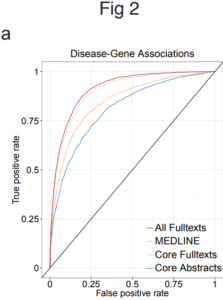
The biggest gain in performance when using full text was seen in finding associations between diseases and genes
The main advantages of text mining full-text scientific articles are volume, information diversity and the inclusion of secondary findings. Unsurprisingly, full-text articles contain more named entities and connections between those entities.
In the case of these 15 million scientific articles, the biggest performance gain in mining full-text articles was the associations found between diseases and genes.
A common mineable format would produce higher quality results
Despite the perceived benefits of mining abstracts mentioned above, bioinformaticians are aware that full-text articles are likely to yield more information and contain more relationships between named entities than abstracts. The problem isn’t lack of text mining awareness; it’s contending with multiple formats and inconsistent licensing terms.
XML is the preferred format used in text mining software. XML is a markup language used to encode documents in a format that is easily read by computers. It is used widely for encoding documents so that computer programs can parse or display the content appropriately.
The study suggests if all articles were available in a structured XML format, it would have “no doubt produced a higher quality corpus.”
In an interview with Science, co-author Lars Juhl Jensen said converting full-text PDF articles into XML formatting is one of the reasons why full-text mining isn’t typically done at scale.
“We probably spent more computational resources teasing the text out of PDFs and beating it into shape than we spent on the actual text mining,” Jensen said.
As information professionals begin to understand the benefits text mining can have across functions – early phase research, pharmacovigilance, IDMP compliance, and more – the desire to find a better way to mine full-text articles will become greater.
At CCC we’ve developed integrated solutions that make it simple to license, access, semantically enrich and index full-text XML articles from a wide range of scientific publishers. Learn more about RightFind XML here.
Keep Learning: